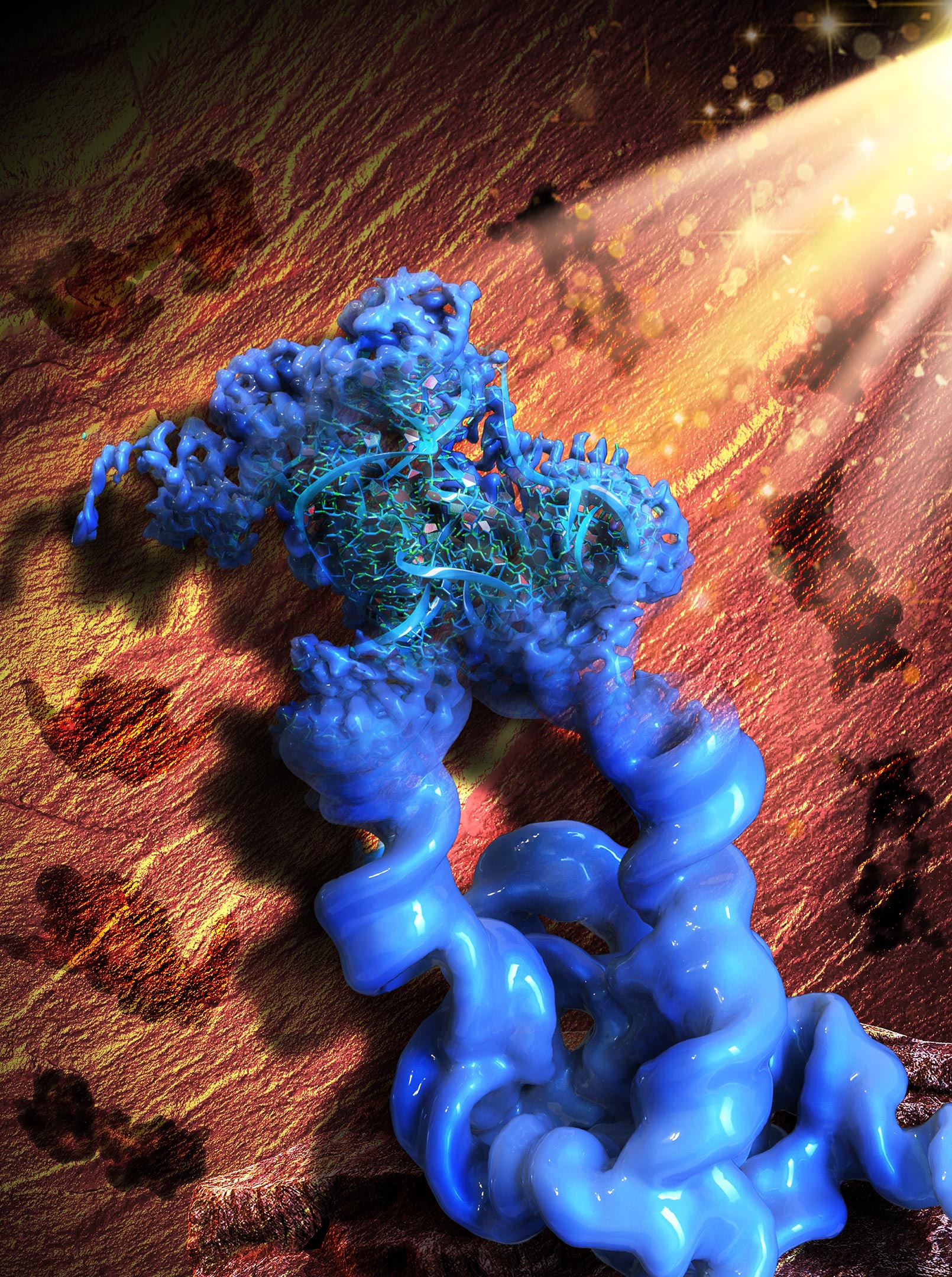
This illustration is impressed by the Paleolithic rock portray in the Lascaux cave, signifying the acronym of our draw, ROCK. Figuratively, the patterns of the rock art in the background (brown) are the 2D projections of the engineered dimeric manufacture of the Tetrahymena neighborhood I intron, whereas the major object in the entrance (blue) is the reconstructed 3D cryo-EM plan of the dimer, with one monomer in point of curiosity and refined to the excessive resolution that allowed the collaborators to manufacture an atomic mannequin of the RNA. Credit: Wyss Institute at Harvard College
Mixture of nucleic acid nanotechnology and cryo-EM provides unparalleled insights into the structures of giant and puny RNAs, advancing RNA biology and drug manufacture.
We dwell in an international created and urge by RNA, the equally crucial sibling of the genetic molecule DNA. In actuality, evolutionary biologists hypothesize that RNA existed and self-replicated even earlier than the appears to be like to be like of DNA. Mercurial forward to current-day other folks: science has printed that lower than 3% of the human genome is transcribed into messenger RNA (mRNA) molecules that in flip are translated into proteins. In difference, 82% of it’s transcribed into RNA molecules with utterly different functions many of which are but unknown.
Ribonucleic acid (RNA) is a polymeric molecule that is awfully crucial in varied biological roles in coding, decoding, legislation and expression of genes. Each RNA and deoxyribonucleic acid (DNA) are nucleic acids. In conjunction with lipids, proteins, and carbohydrates, nucleic acids constitute one of the most four major macromolecules very crucial for all known kinds of life. RNA, like DNA, is assembled as a series of nucleotides, but unlike DNA, RNA is display hide in nature as a single strand folded onto itself, in put apart of a paired double strand.
To fancy what an individual RNA molecule does, its 3D constructing needs to be deciphered at the stage of its constituent atoms and molecular bonds. Researchers have routinely studied DNA and protein molecules by turning them into recurrently packed crystals that is almost definitely examined with an X-ray beam (X-ray crystallography) or radio waves (nuclear magnetic resonance). Alternatively, these ways can no longer be applied to RNA molecules with nearly the equal effectiveness because their molecular composition and structural flexibility dwell them from with out peril forming crystals.
Now, a research collaboration led by Wyss Core College member Peng Yin, Ph.D. at the Wyss Institute for Biologically Impressed Engineering at Harvard College, and Maofu Liao, Ph.D. at Harvard Clinical College (HMS), has reported a fundamentally new technique to the structural investigation of RNA molecules. ROCK, as it’s called, makes exercise of an RNA nanotechnological methodology that lets in it to assemble loads of equal RNA molecules into a extremely organized constructing, which enormously reduces the flexibleness of individual RNA molecules and multiplies their molecular weight. Applied to notorious mannequin RNAs with utterly different sizes and functions as benchmarks, the team confirmed that their draw permits the structural analysis of the contained RNA subunits with a methodology known as cryo-electron microscopy (cryo-EM). Their reach is reported in the journal Nature Strategies.
“ROCK is breaking basically the most popular limits of RNA structural investigations and permits 3D structures of RNA molecules to be unlocked which could well be tough or no longer doable to entry with present programs, and at advance-atomic resolution,” said Yin, who together with Liao led the glimpse. “We ask this reach to invigorate many areas of traditional research and drug pattern, including the burgeoning field of RNA therapeutics.” Yin moreover is a frontrunner of the Wyss Institute’s Molecular Robotics Initiative and Professor in the Department of Systems Biology at HMS.
“ROCK is breaking basically the most popular limits of RNA structural investigations and permits 3D structures of RNA molecules to be unlocked which could well be tough or no longer doable to entry with present programs, and at advance-atomic resolution. We ask this reach to invigorate many areas of traditional research and drug pattern, including the burgeoning field of RNA therapeutics.”
— Peng Yin
Gaining protect a watch on over RNAYin’s team at the Wyss Institute has pioneered varied approaches that enable DNA and RNA molecules to self-assemble into huge structures in step with utterly different tips and requirements, including DNA bricks and DNA origami. They hypothesized that such solutions could well well well moreover be former to assemble naturally occurring RNA molecules into extremely ordered circular complexes wherein their freedom to flex and circulation is extremely restricted by particularly linking them together. Many RNAs fold in complicated but predictable ways, with puny segments horrible-pairing with every utterly different. The result in total is a stabilized “core” and “stem-loops” bulging out into the periphery.
In ROCK (RNA oligomerization-enabled cryo-EM by task of installing kissing loops), a plan RNA is engineered for the self-assembly of a closed homomeric ring by task of kissing-loop sequences (crimson) which could well be put in onto functionally nonessential, peripheral helices (blue). After identifying engineerable nonessential peripheral helices, the length of the helices connecting the kissing-loop motif and the core of the plan RNA is computationally optimized. An RNA manufacture with loads of individual subunits of the plan RNA is transcribed, assembled, after which purified by gel electrophoresis, earlier than it’s a ways almost definitely analyzed by task of the cryo-EM draw. Credit: Wyss Institute at Harvard College
“In our draw we set up ‘kissing loops’ that link utterly different peripheral stem-loops belonging to two copies of an equal RNA in a draw that lets in a overall stabilized ring to be fashioned, containing loads of copies of the RNA of curiosity,” said Di Liu, Ph.D., one of two first-authors and a Postdoctoral Fellow in Yin’s neighborhood. “We speculated that these greater-uncover rings is almost definitely analyzed with excessive resolution by cryo-EM, which had been applied to RNA molecules with first success.”
Picturing stabilized RNAIn cryo-EM, many single particles are flash-frozen at cryogenic temperatures to dwell any further movements, after which visualized with an electron microscope and the relieve of computational algorithms that review the quite a bit of aspects of a particle’s 2D ground projections and reconstruct its 3D architecture. Peng and Liu teamed up with Liao and his previous college graduate student François Thélot, Ph.D., the utterly different co-first creator of the glimpse. Liao alongside with his neighborhood has made crucial contributions to the impulsively advancing cryo-EM field and the experimental and computational analysis of single particles fashioned by particular proteins.
“Cryo-EM has good advantages over frail programs in seeing excessive-resolution info of biological molecules including proteins, DNAs and RNAs, however the puny size and bright tendency of most RNAs dwell winning decision of RNA structures. Our novel draw of assembling RNA multimers solves these two complications at the equal time, by rising the size of RNA and reducing its motion,” said Liao, who moreover is an Affiliate Professor of Cell Biology at HMS. “Our draw has opened the door to rapid constructing decision of many RNAs by cryo-EM.” The combination of RNA nanotechnology and cryo-EM approaches led the team to name their draw “RNA oligomerization-enabled cryo-EM by task of installing kissing loops” (ROCK).
To provide proof-of-precept for ROCK, the team mad by a huge intron RNA from Tetrahymena, a single-celled organism, and a puny intron RNA from Azoarcus, a nitrogen-fixing bacterium, as properly as the so-called FMN riboswitch. Intron RNAs are non-coding RNA sequences scattered in some unspecified time in the future of the sequences of freshly-transcribed RNAs and must be “spliced” out in uncover for the mature RNA to be generated. The FMN riboswitch is display hide in bacterial RNAs serious about the biosynthesis of flavin metabolites derived from food plan B2. Upon binding one of them, flavin mononucleotide (FMN), it switches its 3D conformation and suppresses the synthesis of its mother RNA.
Of their analysis of the Tetrahymena neighborhood I intron, the researchers serene about 105,000 single-particle cryo-EM photos of the ROCK-enabled constructing, and over a series of computational analysis steps reconstructed its constructing, reaching an overall resolution of 2.98 Å, and a resolution of 2.85 Å for the core of the improvement. The final fashions supplied a detailed search for of the Tetrahymena neighborhood I intron, including the previously unknown peripheral domains (proven in brown and crimson), which constitute a belt surrounding the core. Credit: Wyss Institute at Harvard College
“The assembly of the Tetrahymena neighborhood I intron into a ring-like constructing made the samples extra homogenous, and enabled the usage of computational tools leveraging the symmetry of the assembled constructing. Whereas our dataset is pretty modest in size, ROCK’s innate advantages allowed us to solve the improvement at an unparalleled resolution,” said Thélot. “The RNA’s core is resolved at 2.85 Å [one Ångström is one ten-billions (US) of a meter and the preferred metric used by structural biologists], revealing detailed aspects of the nucleotide bases and sugar backbone. I don’t judge shall we have gotten there with out ROCK – or a minimum of no longer with out significantly extra resources.”
Cryo-EM moreover is ready to put off molecules in utterly different states if they, as an illustration, replace their 3D conformation as allotment of their feature. Applying ROCK to the Azoarcus intron RNA and the FMN riboswitch, the team managed to identify the utterly different conformations that the Azoarcus intron transitions through at some stage in its self-splicing direction of, and to suppose the relative conformational rigidity of the ligand-binding put apart of the FMN riboswitch.
“This glimpse by Peng Yin and his collaborators elegantly reveals how RNA nanotechnology can work as an accelerator to reach utterly different disciplines. Being in a position to visualize and understand the structures of many naturally occurring RNA molecules can have huge affect on our realizing of many biological and pathological processes across utterly different cell kinds, tissues, and organisms, and even enable new drug pattern approaches,” said Wyss Founding Director Donald Ingber, M.D., Ph.D., who’s moreover the Judah Folkman Professor of Vascular Biology at Harvard Clinical College and Boston Youngsters’s Sanatorium, and the Hansjörg Wyss Professor of Bioinspired Engineering at the Harvard John A. Paulson College of Engineering and Applied Sciences.
Reference: “Sub-3-Å cryo-EM constructing of RNA enabled by engineered homomeric self-assembly” by Di Liu, François A. Thélot, Joseph A. Piccirilli, Maofu Liao and Peng Yin, 2 Might presumably maybe moreover merely 2022, Nature Strategies.
DOI: 10.1038/s41592-022-01455-w
The glimpse used to be moreover authored by Joseph Piccirilli, Ph.D., an knowledgeable in RNA chemistry and biochemistry and Professor at The College of Chicago. It used to be supported by the Nationwide Science Foundation (NSF; grant# CMMI-1333215, CCMI-1344915, and CBET-1729397), Air Force Situation of industrial of Scientific Learn (AFOSR; grant MURI FATE, #FA9550-15-1-0514), Nationwide Institutes of Health (NIH; grant# 5DP1GM133052, R01GM122797, and R01GM102489), and the Wyss Institute’s Molecular Robotics Initiative.